|
You are here > Biopharmaceutical
Glossaries & Taxonomies Homepage/Search > Informatics >
Bioinformatics > Bioinformatics Information Resources
Bioinformatics
Information Resources
Evolving Terminology for Emerging Technologies
Comments? Questions?
Revisions?
Mary Chitty MSLS mchitty@healthtech.com
Last revised
July 10, 2019
Poster
for BioIT World 2014
Bioinformatics
Information Resources
Mary Chitty, MSLS, Library Director & Taxonomist, Cambridge Healthtech,
Needham MA 02494 USA
Abstract
Biological
data is very noisy and disparate, and reproducible results are elusive. More and
more, researchers and the pharmaceutical industry are turning to powerful
software to interpret genomic, molecular, and clinical information and make
meaningful comparisons. The paradox of “big data” is that, as databases grow
to sizes that are difficult to manage and parse, even more data is needed –
particularly longitudinal data correlating genotypes and phenotypes with
clinical outcomes. In this environment, keeping track of where all the data is,
how to access it, and monitoring data quality and integrity are often
underestimated challenges. Because much useful data is stored as unstructured
text, the interoperability of databases relies on disambiguating drug, gene and
protein names, and mapping ontologies and taxonomies. This poster lists and
annotates valuable resources for tapping the potential of bioinformatics. These
include computational tools for analyzing genomic and molecular data, knowledge
bases where state-of-the-art research in computational biology is collected, and
terminology resources that help diverse databases to communicate with one
another, multiplying their discovery potential.
What is bioinformatics? Clinical
bioinformatics Translational bioinformatics Expert perspectives and definitions
Key Terminology Resources,
Associations, Conferences, Databases, Government Agencies, Journals,
Newsletters, and Software
What
is bioinformatics? Expert
perspectives and definitions
The definition of bioinformatics is not universally agreed
upon. Generally speaking, we define it as the creation and development of
advanced information and computational technologies for problems in biology,
most commonly molecular biology (but increasingly in other areas of biology). As
such, it deals with methods for storing, retrieving and analyzing biological
data, such as nucleic acid (DNA/RNA) and protein sequences, structures,
functions, pathways and genetic interactions. Some
people construe bioinformatics more narrowly, and include only those issues
dealing with the management of genome project sequencing data. Others construe
bioinformatics more broadly and include all areas of computational biology,
including population modeling and numerical simulations. Biomedical
informatics is a slightly broader umbrella that includes not only
bioinformatics, but other areas of informatics in biology, medicine and
health-care. They are closely related. Russ Altman in BIoinformatics
Methods and Applications, 2008
https://books.google.com/books?id=YP1rEFxFgDcC&source=gbs_navlinks_s
Despite the apparent fatigue of the linguistic use of the term
itself, bioinformatics has grown perhaps to a point beyond recognition. We
explore both historical aspects and future trends and argue that as the field
expands, key questions remain unanswered and acquire new meaning while at the
same time the range of applications is widening to cover an ever increasing
number of biological disciplines. Rise and Demise of Bioinformatics? Promise and
Progress, Christos A. Ouzounis, PLOS
Computational Biology April 2012 http://www.ploscompbiol.org/article/info%3Adoi%2F10.1371%2Fjournal.pcbi.1002487
Bioinformatics is the emerging field that deals with the
application of computers to the collection, organization, analysis,
manipulation, presentation, and sharing of biologic data. A central component of
bioinformatics is the study of the best ways to design and operate biologic
databases. This is in contrast with the field of computational biology, where
specific research questions are the primary focus. Bioinformatics, National
Academies Press, 2000 http://www.ncbi.nlm.nih.gov/books/NBK44939/
We have coined the term Bioinformatics for the study of
informatic processes in biotic systems. Our Bioinformatic approach typically
involves spatial, multi- leveled models with many interacting entities whose
behavior is determined by local information. Theoretical Biology Group, Univ. of
Utrecht, Netherlands, Paulien Hogeweg Director http://www-binf.bio.uu.nl/
clinical bioinformatics: Different from other informatics, clinical
bioinformatics should focus more on clinical informatics, including patient
complaints, history, therapies, clinical symptoms and signs, physician's
examinations, biochemical analyses, imaging profiles, pathologies and other
measurements. It was emphasized that the simultaneous evaluation of clinical and
basic research could improve medical care, care provision data, and data
exploitation methods in disease therapy and algorithms for the analysis of such
heterogeneous data sets. Clinical
bioinformatics: a new emerging science Xiangdong Wang
and Lance Liotta, Journal of Clinical Bioinformatics 1:1 2011 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3097094/
Translational
bioinformatics is the development of storage, analytic, and interpretive
methods to optimize the transformation of increasingly voluminous biomedical
data, and genomic data, into proactive, predictive, preventive, and
participatory health. Translational bioinformatics includes research on the
development of novel techniques for the integration of biological and clinical
data and the evolution of clinical informatics methodology to encompass
biological observations. The end product of translational bioinformatics is
newly found knowledge from these integrative efforts that can be disseminated to
a variety of stakeholders, including biomedical scientists, clinicians, and
patients.
AMIA American Medical Informatics Association, Translational bioinformatics http://www.amia.org/applications-informatics/translational-bioinformatics
Growth of the bioinformatics literature: articles
per year 1984-2013
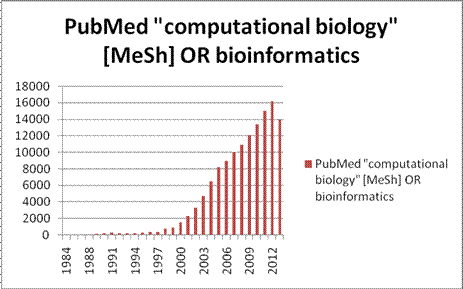 growth
of GenBank total sequences 1982-2013
growth
of GenBank total sequences 1982-2013
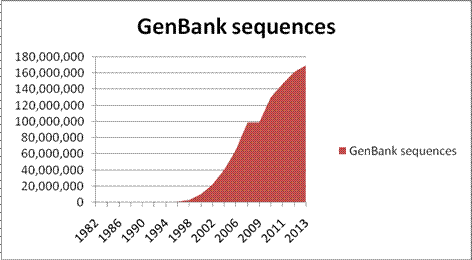
Terminology
Resources
Bioinformatics Glossary, Mary Chitty, Genomic
Glossaries & Taxonomies http://www.genomicglossaries.com/content/Bioinformatics_gloss.asp
Gene Ontology GO http://www.geneontology.org/
Bioinformatics
initiative with the aim of standardizing the representation of gene and gene
product attributes across species and databases. The project provides a
controlled vocabulary of terms for
describing gene product characteristics and gene product annotation data from
GO Consortium members, as well as tools to access and process this data.
HUGO Human Gene Nomenclature Committee http://www.hugo-international.org/comm_genenomenclaturecommittee.php
genenames.org is
a curated online repository of HGNC-approved gene nomenclature, gene families
and associated resources including links to genomic, proteomic and phenotypic
information. HUGO is the Human Genome Organization.
Human Phenotype Ontology http://www.human-phenotype-ontology.org/
Aims to provide a standardized vocabulary of phenotypic
abnormalities.
PubMed MeSH
Medical Subject headings computational biology http://www.ncbi.nlm.nih.gov/mesh/68019295 maps to bioinformatics, and includes
genomics, metabolomics and systems biology.
Associations
bioinformatics.org
http://www.bioinformatics.org/
International Society for Computational Biology http://www.iscb.org/
Founded 1997. Sponsors the journal
Bioinformatics and a number of conferences.
Japanese Society for Bioinformatics http://www.jsbi.org/en/
Conferences
BioIT World http://www.bio-itworldexpo.com/ Enabling
technologies that are driving biomedical research and the drug development
process. 2002-
CSHALS Conference on Semantics in Healthcare and Life Sciences http://lanyrd.com/2014/cshals/
Use
of semantic technologies in the pharmaceutical industry, including
hospitals/healthcare institutions and academic research labs. 2008-
Intelligent
Systems in Molecular Biology http://www.iscb.org/about-ismb
Annual ISCB meeting 1993-
Pacific Symposium on Biocomputing http://psb.stanford.edu/
Current
research in the theory and application of computational methods in problems of
biological significance. 1996-
RECOMB Research in Computational Molecular Biology http://www.recomb.org/home
Scientific
forum for theoretical advances in computational biology and their applications
in molecular biology and medicine. 1997-
Databases
ExPASy,
Swiss Institute of Bioinformatics http://www.expasy.org/
Resource portal, databases, software tools.
Originally Expert Protein Analysis System
International Nucleotide Sequence Database Collaboration http://www.insdc.org/
GenBank, ENA European Nucleotide Archive, DDBJ DNA Databank Japan
Nucleic Acids Research database annual issue http://www.oxfordjournals.org/nar/database/c/
Literature
databases include PubMed, Embase and BIOSIS. Patents databases are also
relevant, as are electronic medical records.
Wikipedia,
Biological databases http://en.wikipedia.org/wiki/List_of_biological_databases
Government
Agencies
Big
data to knowledge BD2K, NIH http://bd2k.nih.gov/#sthash.gptB5T7g.dpbs
BD2K aims to develop the new approaches, standards, methods, tools, software and
competencies that will enhance the use of biomedical Big Data by supporting
research, implementation and training in data science and other relevant fields.
Biomedical
Informatics Research Network http://www.birncommunity.org/
National initiative to advance biomedical research through data sharing
and online collaboration. Funded by the National Institute of General Medicine Sciences (NIGMS).
BISTI
Biomedical Information Science and Technology Initiative http://www.bisti.nih.gov/
Consortium
of representatives from each of the NIH institutes and centers. Established in
2000 to serve as the focus of biomedical computing issues at the NIH.
ELIXIR
http://www.elixir-europe.org/ European life sciences infrastructure for
sharing biological data
Korean Bioinformation Center, KRIBB https://www.kobic.re.kr/newkobic/index.php?mid=eng Core for bioinformatics research.
NCBI National Center for Biotechnology Information, NIH http://www.ncbi.nlm.nih.gov/ National resource for molecular biology
information Part of the National Library of Medicine. Established 1988.
Databases, software and more.
Journals
Bioinformatics, ISCB, Oxford University Press http://bioinformatics.oxfordjournals.org/
BMC Bioinformatics, BioMedCentral http://www.biomedcentral.com/bmcbioinformatics
Briefings
in Bioinformatics, Oxford University Press http://bib.oxfordjournals.org/
Journal
of Clinical Bioinformatics, BioMedCentral http://www.jclinbioinformatics.com/
Molecular Informatics, Wiley http://onlinelibrary.wiley.com/journal/10.1002/(ISSN)1868-1751
PLOS Computational Biology, Public Library of Science http://www.ploscompbiol.org/
Newsletters
Bioinform, Genomeweb http://www.genomeweb.com/newsletter/bioinform
BioIT World http://www.bio-itworld.com/
Application of informatics, IT and computer science in biomedical
research and drug discovery.
Software
NCBI Sequence analysis tools http://www.ncbi.nlm.nih.gov/guide/sequence-analysis/
Includes BLAST Basic Local Alignment Search Tool.
Open Bioinformatics Foundation http://www.open-bio.org/wiki/Main_Page
Projects include BioJava, Bioperl, BioPython, BioRuby, BioSQL, MOBY.
Wikipedia, Bioinformatics software http://en.wikipedia.org/wiki/Category:Bioinformatics_software
Thanks to Aaron Krol, of BioIT World and Clinical Informatics
News for valued editorial advice.
Conclusion:
This is a (very selective) work in progress.
Please let me know of your favorite resources to consider adding.
Mary Chitty mgchitty@healthtech.,com
Bibliography
How
to look for other unfamiliar terms
|
 growth
of GenBank total sequences 1982-2013
growth
of GenBank total sequences 1982-2013
